Section: New Results
Meristem functioning and development
In axis 2 work focuses on the creation of a virtual meristem, at cell resolution, able to integrate the recent results in developmental biology and to simulate the feedback loops between physiology and growth. The approach is subdivided into several sub-areas of research.
Data acquisition and design of meristem models
-
Improvement of the MARS-ALT pipeline robustness.
Meristem, laser microscopy, image reconstruction, cell segmentation, automatic lineaging
Participants : Léo Guignard,, Christophe Godin, Christophe Pradal, Grégoire Malandain [Morpheme, Inria] , Gaël Michelin [Morpheme, IPL Morphogenetics, Inria] , Guillaume Baty, Sophie Ribes [IBC, UM] , Jan Traas [RDP, ENS Lyon] , Patrick Lemaire [CRBM, CNRS] , Yassin Refahi [RDP, ENS-Lyon / Sainsbury Lab, Cambridge, UK] .
This research theme is supported by a PhD FRM grant, Jan Traas's ERC, Inria ADT programme and the Morphogenetics Inria Project Lab.
The MARS-ALT (Multi-Angles Registration and Segmentation - Automatic Lineage Tracking) software pipeline [10] automatically performs a segmentation at cell resolution from 3D or 2D voxel images where the membranes/walls are marked (by a die for example) and makes it possible to follow the lineage of these cells through time.
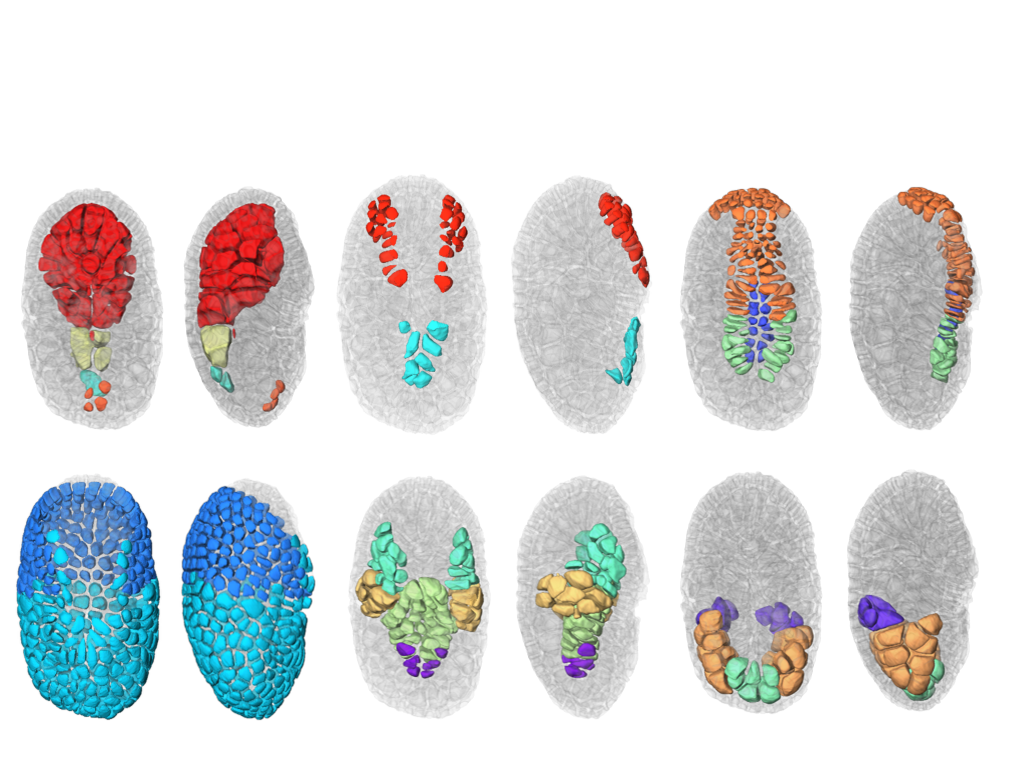
We finalized the development of a a new segmentation and tracking pipeline, ASTEC (Adaptive Segmentation and Tracking of Embryonic Cells). ASTEC is a one-pass algorithm (in contrast to MARS-ALT, that perform first the segmentation and then the tracking in two-passes) that is best suited for movies with numerous close time-points acquired at high spatio-temporal resolution. This pipeline takes advantage of information redundancy across the movies and biological knowledge on the segmented organism to constrain and improve the segmentation and the tracking. We used this one-pass algorithm to segment and track all cell shapes of a developing embryo of the marine invertebrate Phallusia mammillata. As a result we obtained the full track of the shapes of all the cells from the 64 cell stage up to the early tailbud stage (1030 cells undergoing 640 division events followed across 180 time-points through 6 hours of development imaged every 2 minutes, Figure 2). To our knowledge, it is the first time that such high-resolution 4D digital tissues have been generated taking into account the cell shapes.
Based on this quantitative digital representation, we systematically identified cell fate specification events up to the late gastrula stage. Computational simulations revealed that remarkably simple rules integrating measured cell-cell contact areas with spatio-temporal expression data for extracellular signalling molecules are sufficient to explain most early cell inductions. This work suggests that in embryos developing with stereotyped cell shapes and positions (like Phallusia mammillata embryos), the genomic constraints for precise gene expression levels are relaxed, thereby allowing rapid genome evolution. A paper describing the whole approach has been submitted in December 2017.
-
Creating mesh representation of cellular structures.
Participants : Guillaume Cerutti, Sophie Ribes, Christophe Godin, Géraldine Brunoud [RDP, ENS] , Carlos Galvan-Ampudia [RDP, ENS] , Teva Vernoux [RDP, ENS] , Yassin Refahi [RDP, ENS, Sainsbury Lab] .
This research theme is supported the HFSP project Biosensors.
To produce a more efficient data structure accounting for the geometry of cellular tissues, we studied the problem of reconstructing a mesh representation of cells in a complex, multi-layered tissue structure, based either on membrane/wall images segmented using MARS or on nuclei images of shoot apical meristems. The construction of such mesh structures for plant tissues is currently a missing step in the existing image analysis pipelines.
We developed tools to reconstruct a 3D cell complex representing the tissue, based on the dual simplicial complex of cell adjacencies. This set of tetrahedra is optimized from a reasonable initial guess to match the adjacencies in the tissue, which proved to produce a very faithful reconstruction [55]. We also developed a set of methods to triangulate such reconstructions, and enhance the quality of triangular mesh representations of plant tissue, simultaneously along several criteria [54].
These tools are integrated in the DRACO-STEM computational pipeline released as an independent package to enable biomechanical simulations on real-world data.
-
Design of 3D digital atlases of tissue development.
Participants : Sophie Ribes, Yassin Refahi [RDP, ENS, Sainsbury Lab] , Guillaume Cerutti, Christophe Godin, Christophe Pradal, Frédéric Boudon, Gregoire Malandain [RDP, ENS] , Gaël Michelin [RDP, ENS] , Jan Traas [RDP, ENS] , Teva Vernoux [RDP, ENS] , Patrick Lemaire [CRBM, CNRS] .
This research theme is supported the Inria Project Lab Morphogenetics, the ADT Mars-Alt and the HFSP project Biosensors.
To organize the various genetic, physiological, physical, temporal and positional informations, we build a spatialized and dynamic database [61]. This database makes it possible to store all the collected information on a virtual 3D structure representing a typical organ. Each piece of information has to be located spatially and temporally in the database. Tools to visually retrieve and manipulate the information, quantitatively through space and time are being developed. For this, the 3D structure of a typical organ has been created at the different stages of development of the flower bud. This virtual structure contains spatial and temporal information on mean cell numbers, cell size, cell lineages, possible cell polarization (transporters, microtubules), and gene expression patterns. Such 3D digital atlas is mainly descriptive. However, like for classical databases, specific tools make it possible to explore the digital atlas according to main index keys, in particular spatial and temporal keys. Both a dedicated language and a 3D user interface are being designed to investigate and query the 3D virtual atlas. Current developments of this tool consist in using directly the segmented images produced from laser microscopy to build the atlas. To better represent the development of a biological population, a method to compute an "average" structure is being investigated (a manuscript is in preparation).
Shape analysis of meristems
Participants : Jonathan Legrand, Guillaume Cerutti, Pierre Fernique, Frédéric Boudon, Yann Guédon, Christophe Godin, Pradeep Das [RDP, ENS] , Arezki Boudaoud [RDP, ENS] .
The MARS-ALT pipeline provides rich spatio-temporal data sets for analyzing the development of meristems, since it allows to performs 3D cell-segmentation and to compute cell-lineage. This enable the extraction and study of spatio-temporal properties of a tissue at cellular scale. To facilitate the analysis and to structure the obtained data have implemented a dedicated temporal graph structure. In this graph, vertex are cells and edges are spatial or temporal relationships, thus proposing a natural representation of the growing tissue. Various variables can be attached either to the vertices (e.g. cell volume, inertia axes) or the edges (e.g. wall surface, distance between cell centroids). This graph may be augmented by new variables resulting from various spatial or temporal filtering (e.g. cell volumetric growth). Looking at homogeneous regions in the variable space, cellular patterns can be identified, by clustering methods for instance.
Considering the highly-structured nature of our data (time and space structuring) and the potential diversity and heterogeneity of possible cell descriptors, we developed two complementary approaches:
-
A first one that favours the spatial structuring: In this approach, the cell neighbourhood and the cell descriptors are jointly taken into account in a clustering approach whose objective is to identify a small number of clusters corresponding to well-defined cell identities. Once the cells have been labelled using the clustering algorithm, cell generation distributions may be estimated on the basis of the labelled lineage trees.
-
A second one that favours the temporal structuring: In this approach, the data of interest are lineage forest and the only spatial structuring taken into account corresponds to siblings with respect to a given parent cell. In a first step, cell identities are inferred on the basis of the cell descriptors taking into account lineage relationships using hidden Markov tree models and the spatial regions that emerge from the cell identity labelling are then characterized. This second approach is supported by the fact that cell topology is only affected by division which makes highly relevant the local spatial information taken into account in this approach.
Mechanical models of plant tissues
Participants : Olivier Ali, Hadrien Oliveri, Christophe Godin, Jan Traas [ENS-Lyon] .
This research theme was supported, between 2012 and 2017, by the Inria Project Lab Morphogenetics and Jan Traas's ERC.
During the previous years, we set up a multi-scale mechanical model of a growing shoot apical meristem (the specific tissue at the very tip of plants where stem cells are active and produce new organs such as branches, leafs and flowers) with sub-cellular resolution, a detailed description of the core elements of this modelling approach has been developped in our previous reports. The aim of this project is to provide a computational framework for simulating growth of multicellular plant tissue. Several papers (and a reivew) have been published over the past few years on this work, in close collaboration with biologists: [51], [3], [26], [2].
Last year, our simulations pointed out that cell wall remodelling and growth initiation have to be co-regulated in order to initiate young organs formation. Biologists unraveled a biochemical signaling pathway that could explain this synergy. This joint work is currently under submission in a high impact factor journal.
Two years ago, we started to work on the integration of a feedback loop between mechanical stresses and growth (PhD work of Hadrien Oliveri started in Oct. 2015). A close study of this feedback mechanism made us refine several aspects of our modelling approach (tensor formalism to quantify cell polarity, ...). Ever since, Hadrien Oliveri has been studying the influence of this feedback loop on the morphogenesis of an epithelium. FEM-based simulations have been carried out on simple structures as proof of concept. This first step of the work is currently being submitted. This year, we also started to study the influence of such a mechanical-based feedback mechanism on the morphogenesis of real tissues. The specific question we want to investigate concerns flatness: How can plants produce flat organs such as leaves of sepals ? We investigate this question in the context of the sepal formation, always in close collaboration with biologists doing experiments on the very same topic.
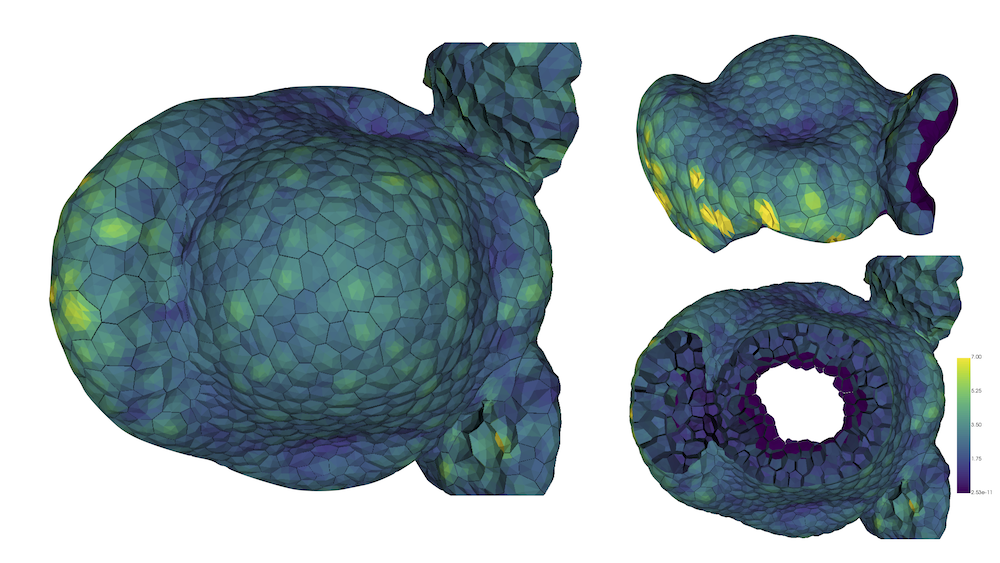
This year, we also started in investigate to a quantitative manner the mechanical influence of inner tissues in the morphogenesis process. Indeed, up to now, our modelling approach was focused on the mechanics of the epidermis, known to be the main load-bearing layer. However, new experimental evidence suggest that inner tissues may influence and/or trigger morphogenesis processess. In order to investigate such mechanisms, our strategy relies on the use of high quality digitized tissues in 3D. Such structures, composed of triangular meshes, are produced through a workflow based on the updated version of the MARS pipeline and the DRACO-STEM module, developped within the team. Currently, numerical simulations are being carried out to analyse the mechanical equilibrium of the structures loaded with pressurized forces, see 4. Preliminary results tend to confirm the leading mechanical role of the epidermis. Interestingly, sharp differences in the mechanical characteristics emerge between epidermal cells and inner ones.
Mechanical modelling of embryo morphogenesis.
Participants : Bruno Leggio, Emmanuel Faure, Patrick Lemaire [CRBM, CNRS] , Christophe Godin.
A work on data analysis and modelling of morphogenesis and development in embryos of ascidians has been started this year. It comprises two main branches: starting from segmented data at cellular resolution, global and local symmetries of embryo development were analyzed. An analysis in terms of entropy of conserved embryonic properties was developed in order to characterise different stages of development as well as different tissues. In parallel, a mechanical and topological analysis of cell-cell interactions was carried out. This lead us to develop a new and original physical model of cleavage-plane determination in different tissues, with the goal of understanding the role of purely mechanical interactions in shaping ascidian embryos.
Gene regulatory networks: Design of a genetic model of inflorescence development.
Participants : Eugenio Azpeitia, Christophe Godin, François Parcy, Etienne Farcot.
This research theme is supported by the Inria Project Lab Morphogenetics.
Modeling gene activities within cells is of primary importance since cell identities correspond to stable combination of gene expression. We studied the regulatory network that controls the flowering transition during morphogenesis. To overcome the network complexity and integrate this regulation during ontogenesis, we have developed a first model of the control of floral initiation by genes, and in particular the situation of cauliflower mutants, in which the meristem repeatedly fails in making a complete transition to the flower. The network was validated by multiple analyses, including sensibility analyses, stable state analysis, mutant analysis, among others. Once the network was validated, it was coupled with an architectural model of plant development using L-systems. The coupled model was used to study how does changes in gene dynamics and expression could change the architectural properties of plants and produce cauliflowers instead of flowers. Finally, the architectural model without the network was used to study how changes in certain parameters could generate different curd morphologies, including the normal cauliflower and the romanesco one.
We have three main types of predictions. (1) How does gene expression is modified from WT to cauliflower organisms. (2) How gene regulate plants shape in order to produce curds instead of flowers. (3) The main parameter regulating curds shapes. The predictions made using the model are currently being experimentally tested. All our results could provide a comprehensive understanding of how does genes and plant architecture are linked in a dynamical way.
Model integration
Participants : Frédéric Boudon, Christophe Godin, Guillaume Cerutti, Jean-Louis Dinh, Eugenio Azpeitia, Jan Traas.
This research theme is supported by the Morphogenetics Inria Project Lab.
One key aspect of our approach is the development of a computer platform dedicated to programming virtual tissue development, TissueLab. This platform, based on OpenAlea, will be used to carry out integration of the different models developed in this research axis. In the past year, progress has been made in defining a generic tissue data structure that would be visualized, manipulated and updated through this platform. Currently, robust geometric operations such as division are implemented and tested. Moreover, a redesign of the structure based on more elaborated formalisms such as combinatorial maps is being investigated. A 2D version is being developed in the context of Jean-Louis's Dinh PhD thesis, and will be described in a forthcoming book chapter.
Our approach consists of building a programmable tissue which is able to accept different modeling components. This includes a central data structure representing the tissue in either 2-D or 3-D, which is able to grow in time, models of gene activity and regulation, models of signal exchange (physical and chemical) between cells and models of cell cycle (which includes cell division). An introduction to the modeling of some main components of such integrated system was published as a book chapter in the series of Ecole de Physique des Houches [12]. For each modeling component, one or several approaches are investigated in depth, possibly at different temporal and spatial scales, using the data available from the partners (imaging, gene networks, and expression patterns). Approaches are compared and assessed on the same data. The objective of each submodel component will be to provide plugin components, corresponding to simplified versions of their models if necessary, that can be injected in the programmable tissue platform. This work is developed in collaboration with the RDP group at ENS-Lyon [64] and the CPIB group in Nottingham, UK [49].